Current Grants
Despite important advances in the treatment of heart failure (HF), >50% of patients die within 5 years of diagnosis at their first hospital admission, and HF remains a leading cause of morbidity, mortality and healthcare expenditure in the United States. With the prevalence of HF expected to increase to 46% by 2030, novel mechanistic insight into HF pathogenesis and strategies to interrupt this progression are a large unmet clinical need. The research in this project focuses on the role of exosomes or extracellular vesicles (EVs) and their cargo RNAs (EV-RNAs) as novel functional biomarkers and as discovery tools for novel pathways within cells. We have discovered and validated plasma RNA signatures that correlate with human HF phenotypes. Importantly, many of these EV-RNAs change in parallel in cardiac tissue, modulating complex signaling pathways that may underlie HF pathogenesis. This work affords a unique opportunity to develop i) novel clinically useful biomarkers for improved risk stratification of HF patients; and ii) novel therapeutic targets to interrupt the adverse remodeling process.
This project broadly addresses the following:
1. Improve the performance (including prognostic/predictive accuracy and coefficient of variance) of plasma RNA biomarkers by more specifically measuring EV-RNAs on validated platforms in biorepository plasma samples from carefully-phenotyped HF and post-MI patients.
2. Determine a functional role for EVs isolated from human HF samples with varied phenotypes in simplified cell culture (iPSC-derived CMs) and organ-on-chip models
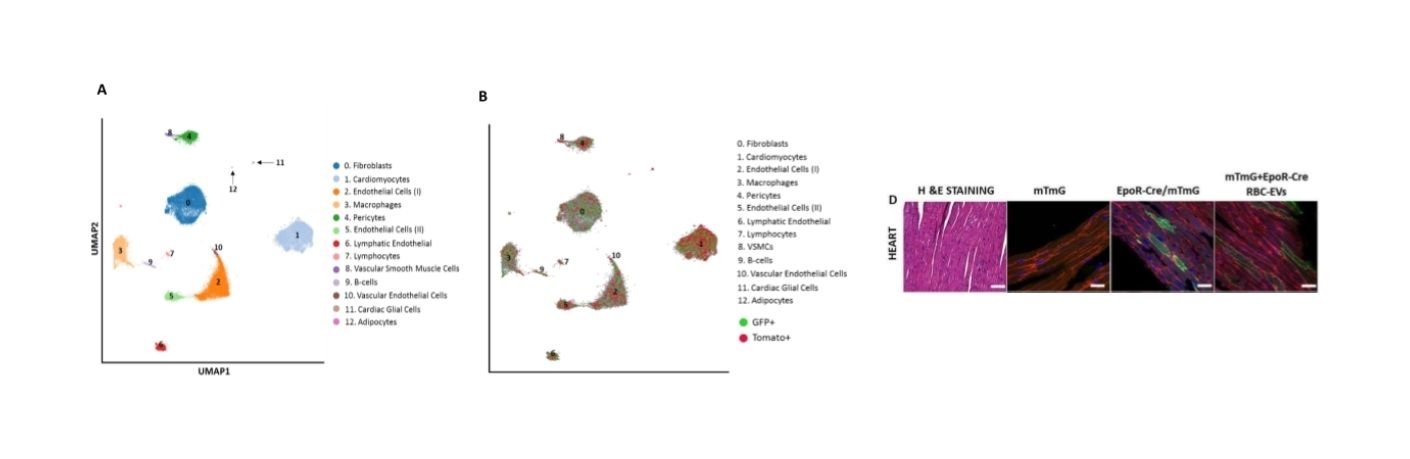
3. Leverage a novel murine model of exosome tracking (ExoMap) mouse to determine the functional consequences of exosome targeting in cardiomyocytes and other cardiac cells in murine models of ischemic and non-ischemic HF using single cell nuclear RNAseq.
4. Identify small molecule regulators of EV release/uptake to manipulate EV-mediated signaling in these murine models.
5. Leverage newly identified cellular RNA biomarkers to develop novel conditional siRNA therapeutics that target cardiac hypertrophy, autophagy and fibrosis.

Publications
TRACE-seq: A transgenic system for unbiased and non-invasive transcriptome profiling of living cells
Circulating miRNAs and Risk of Sudden Death in Patients With Coronary Heart Disease
Extracellular vesicles (EVs) and the RNAs contained within them (EV-RNAs) are secreted into biofluids by every cell type and have emerged as potential prognostic or predictive biomarkers of a wide range of diseases, providing a targetable, accurate real-time representation of the disease state. However, advancement of EV-RNAs in the clinic as biomarkers of disease has been impeded by challenges in their isolation and analysis, most notably the lack of tools and techniques to i) isolate and precisely characterize tissue-specific EV-RNA populations; ii) define heterogeneity in surface markers and RNA content in tissue-specific EV populations; and ii) determine changes in EV-RNAs associated with disease state. In this proposal, we use a collaborative and innovative approach to advance technology that would allow isolation and granular characterization of EV populations from hematopoietic cells, brain, and heart.
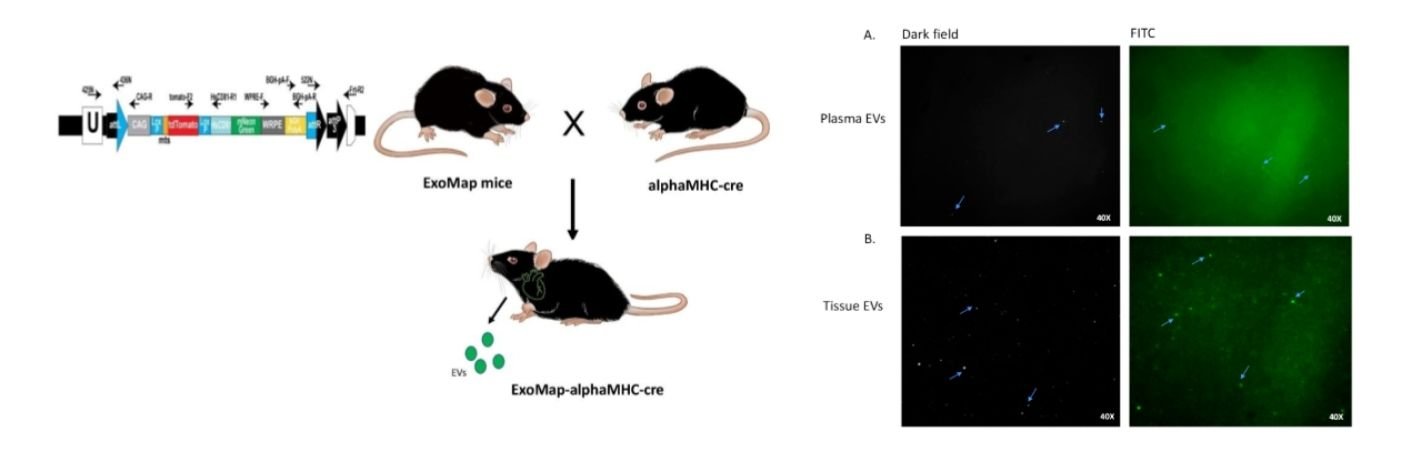
Our objective is to identify cell/tissue specific markers for isolation of EVs using computational analysis, transcriptomics, and EV-tracking in genetic mouse models that allow for fluorescence/antibody-based identification of EVs in a cell-specific manner. Information will also be obtained on individual EVs with a novel quantitative single molecule localization microscopy (qSMLM) approach. qSMLM, a sensitive fluorescence-based imaging method, will be used to quantify the number of affinity isolated EVs, their size, and key RNA content using molecular beacons. The identified tissue-specific EV-markers and EV-RNAs will be used for validation in a variety of human models. Our objective is to determine cellular/tissue contribution to EV-RNAs from tissue-on-chip effluents; and assess dynamic changes in EVs from human plasma from subjects with acute disease (coronary ischemia or cerebrovascular accident) or physiological processes (exercise). Notably, proposed experiments will help determine contribution of different tissues to the plasma biofluid RNA landscape at baseline, and in response to physiological or disease stressors.
Together, the tools and techniques developed in the UG3 phase, and validated in the UH3 phase would serve as a road-map for the discovery and development of EV markers and EV-RNAs specific to other tissues. Ultimately, the use of tissue-specific EV-RNAs to probe disease state would provide a dynamic window into disease progression or regression with higher sensitivity and fidelity compared to currently available technologies.
Publications
A Novel Circulating MicroRNA for the Detection of Acute Myocarditis
Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation
Plasma Circulating Extracellular RNAs in Left Ventricular Remodeling Post-Myocardial Infarction
In this project we examine the dynamics of EV release from cardiomyocytes and other cardiac resident cells, especially resident macrophages in the setting of acute hypoxemia/ischemia. We leverage the ExoMap mouse model that allows for identification and profiling of tissue-specific EVs in mouse models of acute ischemia to study how the characteristics and RNA cargo of cell-specific EVs change with these stressors. TO study these EVs and their contents we have developed tools to specifically isolate EVs from heart tissue and plasma. We have also adopted methodology to study novel small RNAs (tRNA fragments) and discovered novel signatures in human subjects on cardiopulmonary bypass (equivalent of models of nutritional depravation from ischemia and oxidative stress).

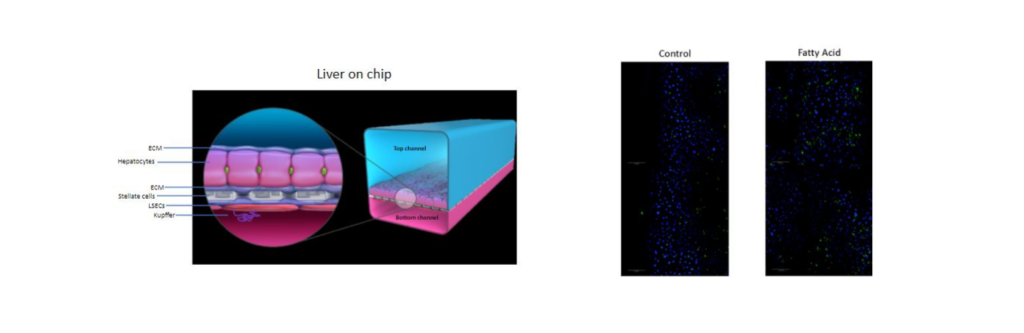
Recent findings suggest that fat tissue can signal to other organs, including the liver and heart in diabetes. These signals play an important role in many diseases that are associated with diabetes and obesity, including abnormal metabolism, heart failure and arrhythmias. We have shown that fat tissue release packets called extracellular vesicles that contain many types of signaling molecules. These packets can travel in the blood to many different organs to alter their function and may be important in the diseases related to obesity. Finally, the vesicles released from fat in the abdomen may be different in their function compared to those released from fat under the skin. Importantly, weight loss surgery can improve the diseases related to obesity and diabetes. In this study we aim to understand the contents of these vesicles from the different types of fat obtained from patients undergoing weight loss surgeries. In addition, we plan to understand how they signal by applying them to heart cells and liver organ-on-chip models and studying changes in the function and transriptome of these cells. Finally, we think that these vesicles from fat are present in blood. We will therefore study how the content of these vesicles changes after weight loss surgery and how that is related to improvement in body fat, heart and liver function.
Contact us
Please feel free to contact us for any inquiries.
Be a Part of Das Lab
We are searching for talented instructors, research fellows, research assistants, and clinical research coordinators. We will be very happy to have you on board.
